Variability of Nonstationary Time Series
In most of the chapter we’ve been talking about the mean trend of a time series and various methods of detecting and removing it. However, there’s another type of nonstationarity where the variability of a time series change with the mean level over time.
Recall that in linear regression we apply a log-transformation on $y$ when the variance of the residuals increases as $\hat{y}$ increases. Similar techniques can be applied for time series data. Here we’re talking about global changes – if the variance changes locally, then a transformation wouldn’t help and different methods would be needed to stabilize the variance.
Log transformation
We’ll use the AirPassengers dataset in R to show how the variability of a time series could be stabilized.
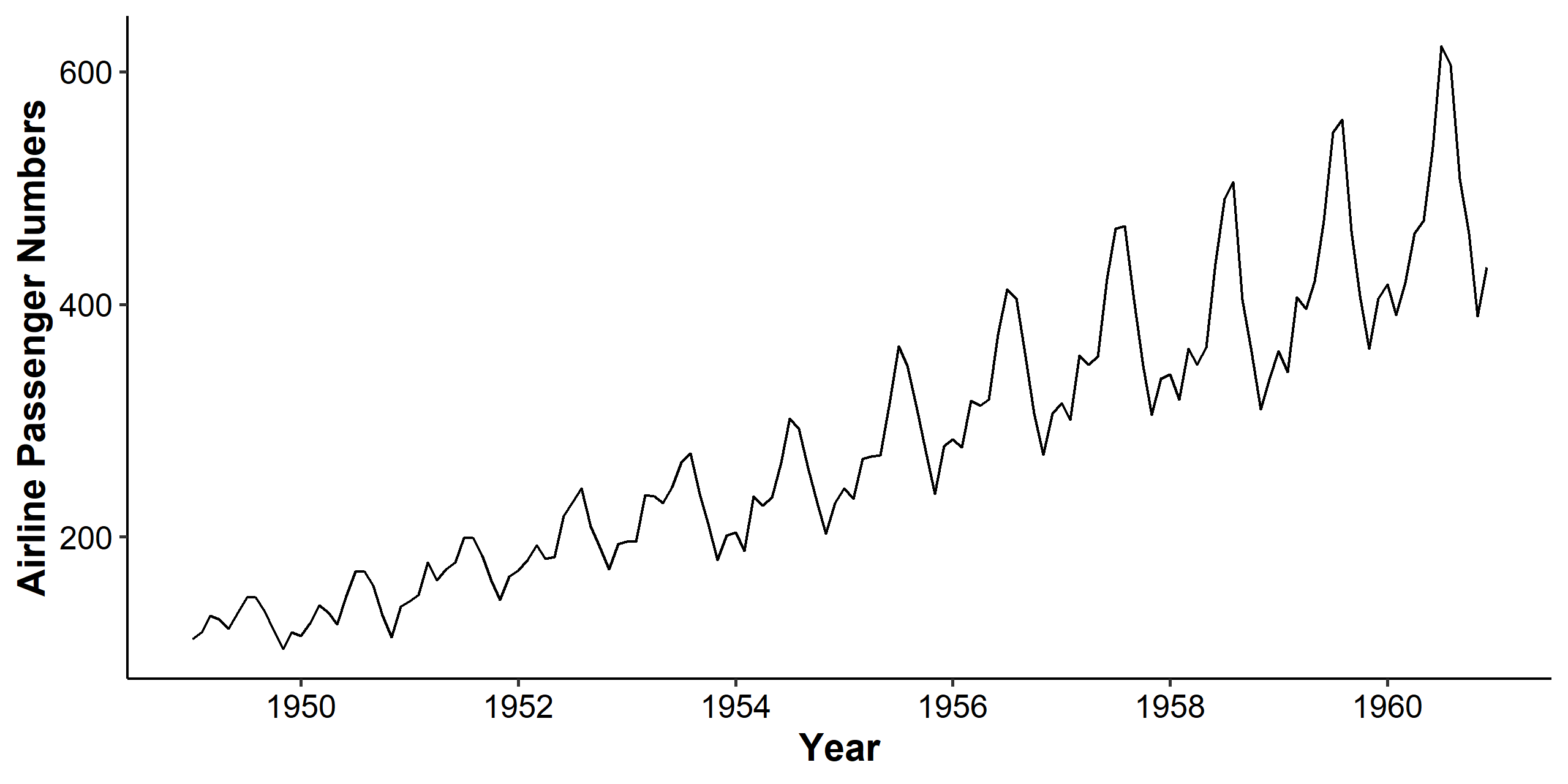
We can see clearly from the time series plot that the mean and variance both increase over time, and there’s a seasonal pattern involved. The increasing mean trend can be handled by differencing, and the seasonal pattern will be discussed in the next chapter.
For stabilizing the variance, we can log-transform the dataset. As we can see below, the mean still increases, but the variance becomes stable across the years.
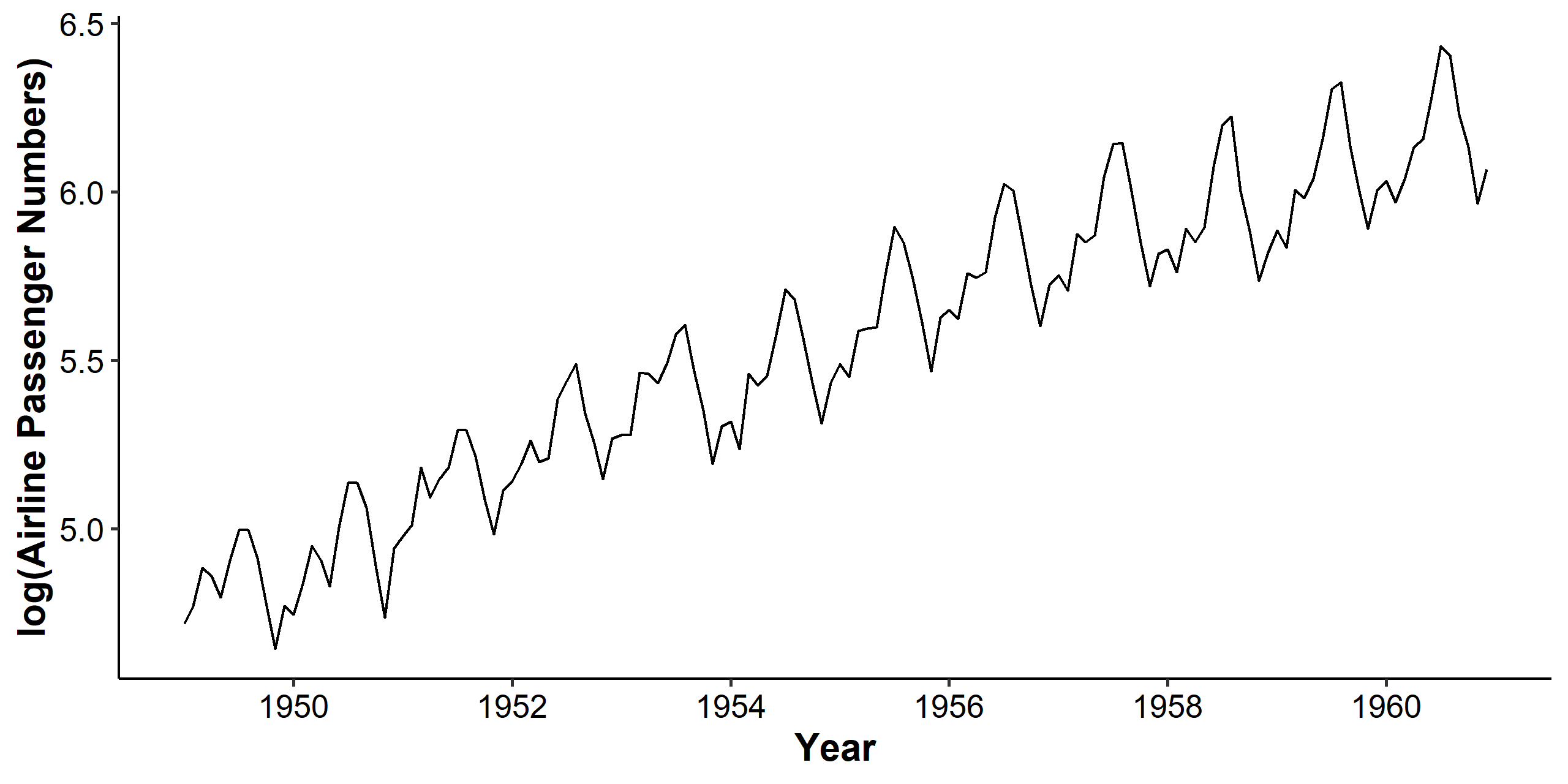
Log-transformation works under the following assumption:
$$ E(X_t) = \mu_t, \quad Var(X_t) = \mu_t^2 \sigma^2 $$
To demonstrate this we are going to need a Taylor expansion of a smooth function $f(x)$ at $x_0$. We will only use the first order approximation of $f(x)$:
$$ f(x) = f(x_0) + f’(x_0)(x - x_0) + \frac{f’’(x_0)}{2!}(x - x_0)^2 + \cdots $$
Now we plug in $x_0 = \mu_t$.
$$ \begin{gathered} \log X_t \approx \log \mu_t + \frac{1}{\mu_t}(X_t - \mu_t) \\ E(\log X_t) \approx E(\log \mu_t) + \frac{1}{\mu_t}E(X_t - \mu_t) = \log \mu_t \\ Var(\log X_t) \approx \frac{1}{\mu_t^2}Var(X_t - \mu_t) = \frac{\mu_t^2\sigma^2}{\mu_t^2} = \sigma^2 \end{gathered} $$
So the variance becomes constant after taking the log-transform. We usually apply log-transformation when the variance of the series increases over time. After log-transformation, we can remove the remaining linear trend through differencing. We’ll discuss seasonal patterns in the next chapter.
Box-Cox power transformation
The log-transformation is a special case of a broader class called the Box-Cox power transformation. The Box-Cox transform is given by
$$ Y_t = \frac{X_t^\lambda - 1}{\lambda} $$
We apply different transformations when $\lambda$ takes different values:
$$ \begin{gathered} \lambda = -1 \Longrightarrow Y_t = \frac{1}{X_t} \\ \lambda = -0.5 \Longrightarrow Y_t = \frac{1}{\sqrt{X_t}} \\ \lambda = 0 \Longrightarrow Y_t = \log X_t \\ \lambda = 0.5 \Longrightarrow Y_t = \sqrt{X_t} \\ \lambda = 1 \Longrightarrow Y_t = X_t \end{gathered} $$
As indicated above, this method requires the time series $X_t$ to be positive. If there exists negative values, a certain constant can be added. In R, we can use FitAR::BoxCox() to find the MLE and a 95% confidence interval for $\lambda$.
Example of sunspot dataset
We’ll apply what we discussed to the sunspot dataset. The dataset contains 289 observations of yearly numbers of sunspots from 1700 to 1988 (rounded to one digit).
Time series plot
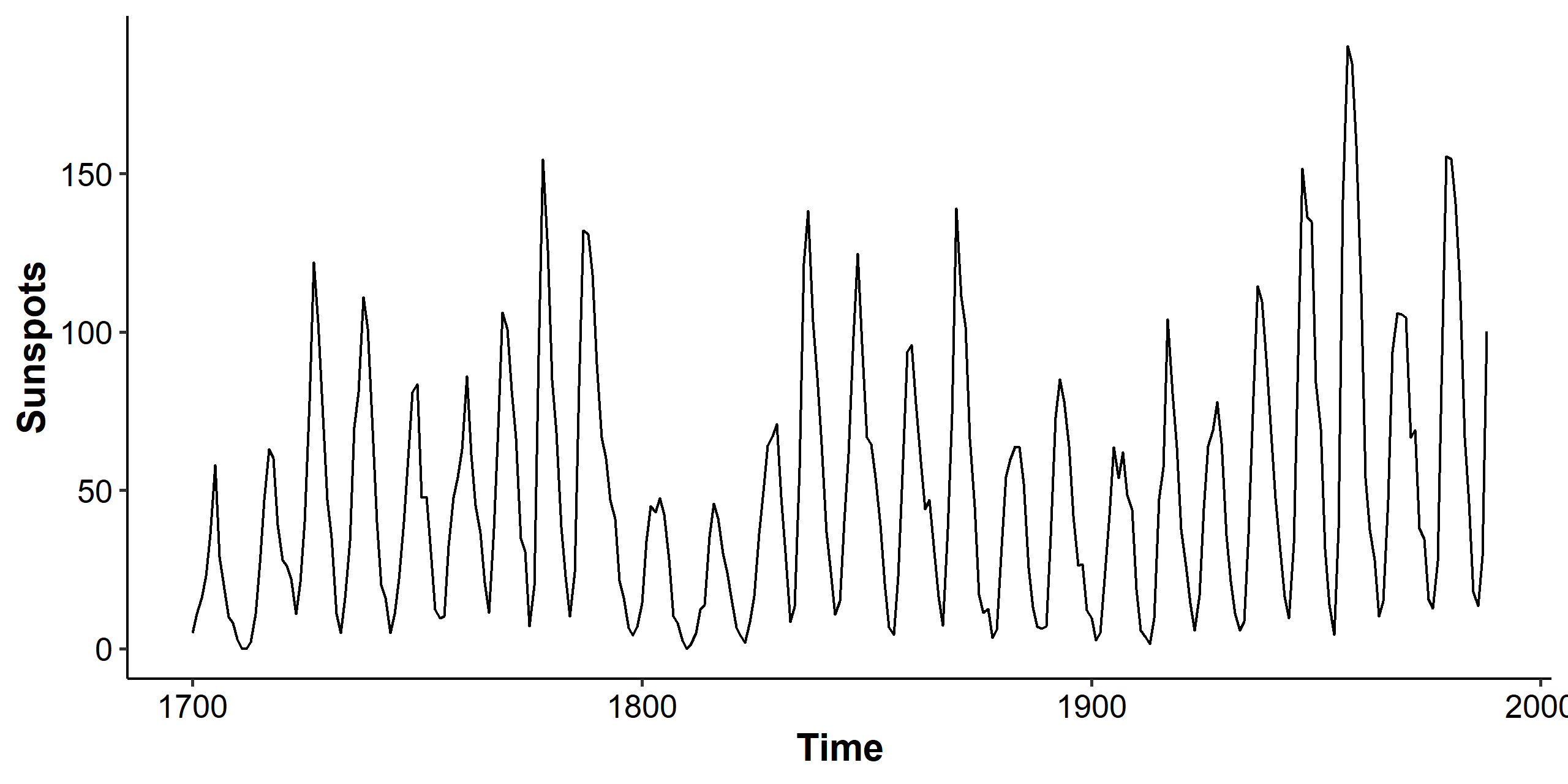
The mean looks somewhat stationary, but the variance is clearly changing over time, so we consider the Box-Cox transformation.
Transformation and differencing
| |
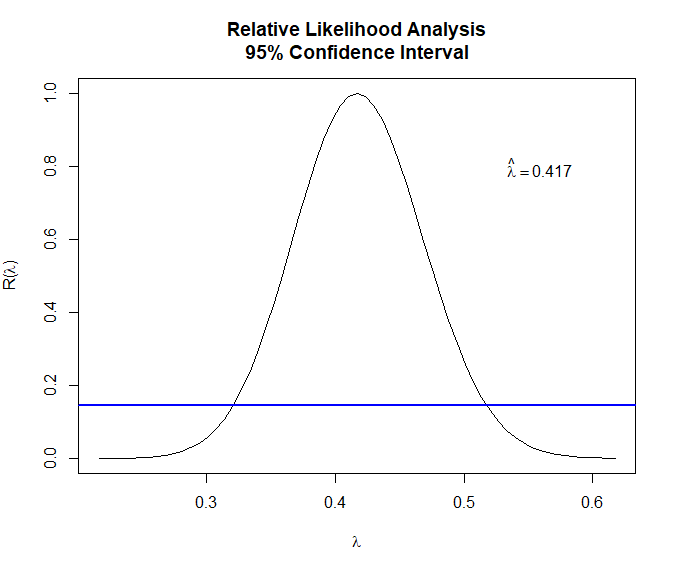
The MLE is $\hat\lambda = 0.417$ and the 95% CI covers 0.35-0.5. We may try $\lambda = 0.5$ and take the square root of the original series so it’s easier to interpret1.
| |
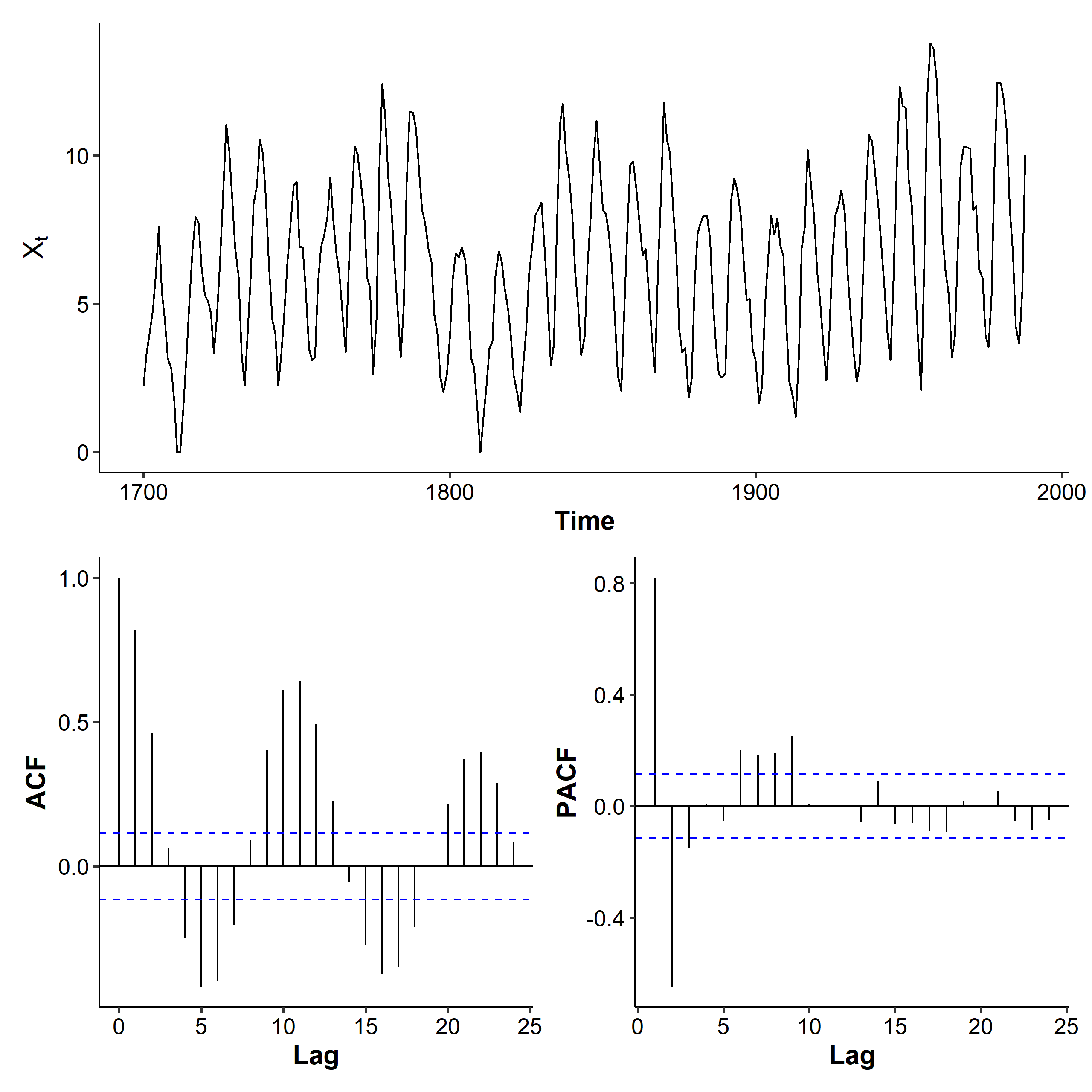
The variance of the transformed series is a lot stabler, although at some points (e.g. year 1800) it’s still not constant. The ACF decreases with a sine wave pattern, and the PACF has spikes at lags 1, 2, and up[ to lag 9. AR(9) is a candidate for this dataset.
We can use the ADF test to check if we need differencing. The p-value is smaller than 0.01, so we reject the null and treat the transformed series as stationary, thus no differencing is needed.
| |
Train-test split and model fitting
We’re going to use the transformed data from now on. A 90-10 split is applied to prepare for calculating the MAPE later. The ar() function can be used to fit an AR model and check the coefficients. We set the order.max parameter as 10 because the last spike in the PACF was at lag 9.
| |
Among the 10 different AR orders, the ar() function selected order 9 as the best model based on its lowest AIC. Note that since some of the coefficients are very small, there’s room for improvement by setting some of those to zero in our model.
It’s obvious from the time series plot that the mean is not zero, but we can use a t-test to make sure:
| |
So we will include the intercept term. Now we can estimate the parameters in the AR(9) model:
| |
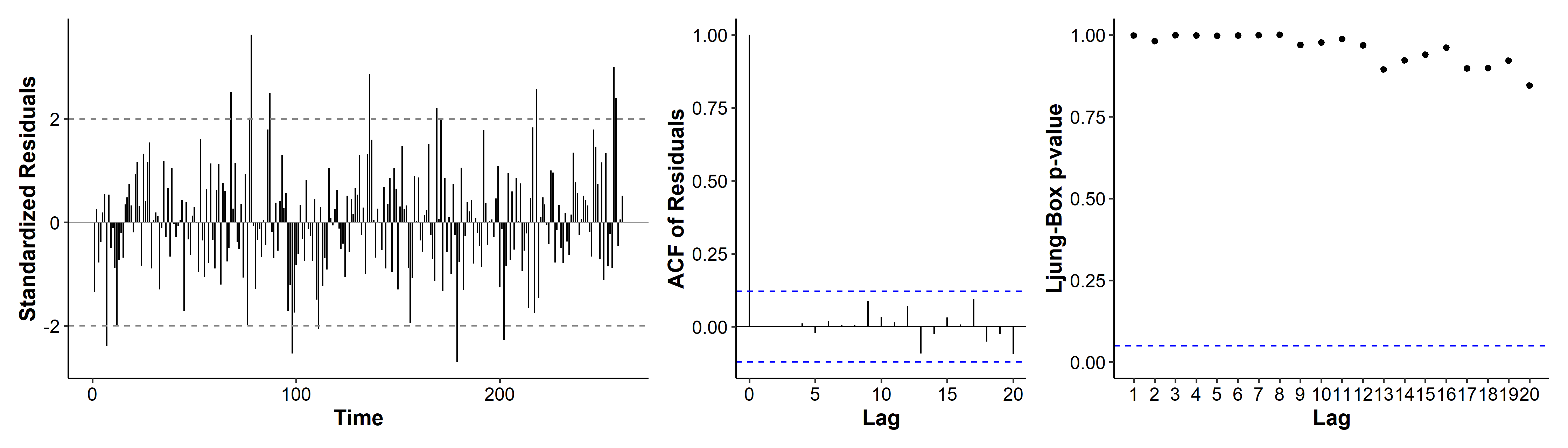
In the diagnostic plots, quite a few standardized residuals are over the $\pm 2$ range. the ACF only has a spike at lag 0, indicating no autocorrelation in the residuals. All p-values for the Ljung-Box tests are very high. Overall there doesn’t seem to be major violations of our model assumptions.
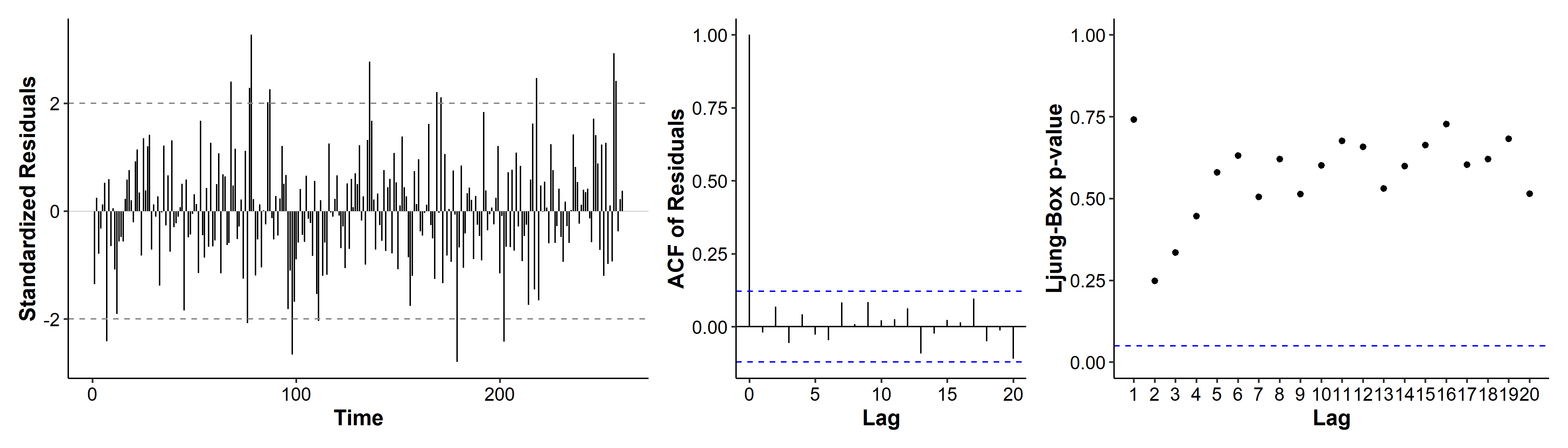
As we said earlier, we can try to reduce the number of parameters in our model. By looking and the coefficients and their standard errors, we may eliminate some parameters. For example, the ar3 coefficient’s CI includes zero, so we may drop it from our model. The same goes for ar6, ar7 and ar8.
| |
We’ve gone from 10 coefficients (9 + intercept) to 6. The diagnosis plots are not too different, and the model assumptions are still met.
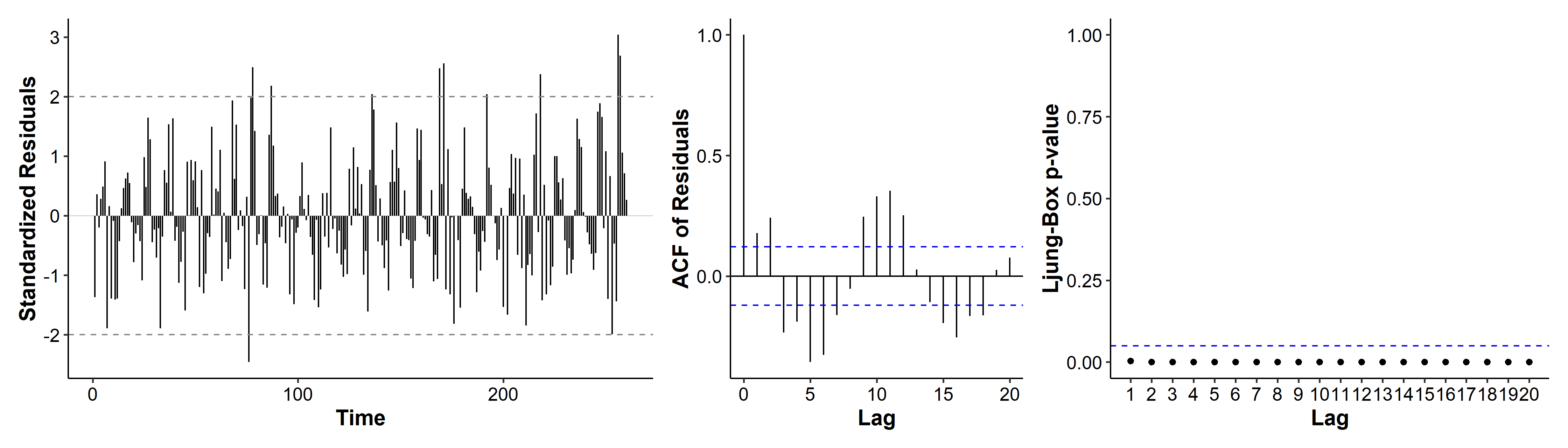
A third model we may consider is ARMA(1, 1), if we consider the patterns in Figure 5 to be tail-off.
| |
The AIC is a lot higher than before! if we look at the diagnostic plots, the ACF has multiple spikes, and the p-values are all below 0.05. ARMA(1, 1) is thus not a good candidate model.
Model selection
To further compare the other two models, we can check their AIC, BIC, number of parameters, and MAPE. The reduced model performs better in terms of all four criteria.
| Criteria | AR(9) | Reduced |
|---|---|---|
| AIC | 783.9829 | 781.3608 |
| BIC | 823.1504 | 806.2856 |
| # parameters | 10 | 6 |
| MAPE | 0.1671 | 0.1657 |
We then fit the entire dataset and get our final model:
$$ (1 - 1.2278B +0.5479B^2 - 0.1132B^4 +0.1149B^5 - 0.2154B^9)(\sqrt{X_t} - 6.3985) = Z_t $$
with $\hat\sigma^2 = 1.116$. We can use this final model to predict future values. Note that the y-axis is still transformed, and we can take the square to visualize the original scale. We can also simplify the plotting code using the plot(forecast(model, 20)) function from the R package forecast. The function also takes a lambda parameter that could automatically transform the series back to its original scale.
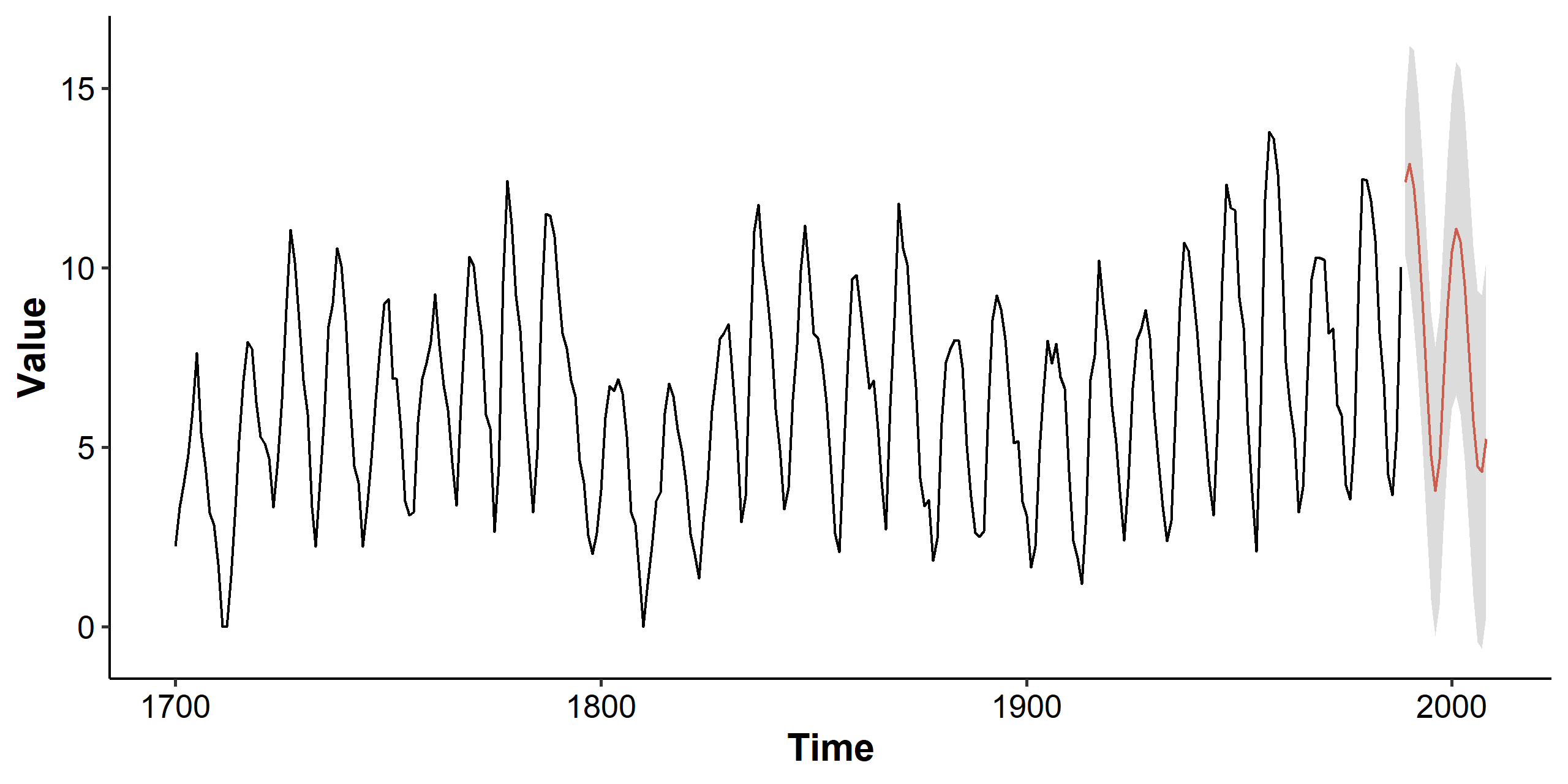
Summary
The general procedure for model fitting is as follows.
- Time series plot. We want to first check for mean and variance changes, as well as seasonal or other patterns.
- Transformation. Using Box-Cox we can determine if transformation is necessary for our data. If it is then we use the transformed data from this step onwards.
- Divide the data. Apply a train-test split on the (transformed) data so that we can calculate the MAPE (or other metrics) for our models. The following steps should only use the training set.
- Check whether differencing is needed. Using the ts plot, ACF and the ADF test to determine if we need to difference the data.
- Determine $p$ and $q$ for the ARIMA model. Use patterns in the ACF and PACF to find candidate models.
- Fit models. In model fitting we should use un-differenced data, because ARIMA will take care of the differencing.
- Model diagnosis. A candidate model shouldn’t have any of its assumptions violated.
- Model selection. Use the AIC, BIC, parameter number and MAPE to find the best model. The testing set will be used in this step.
- Build final model. Use the entire dataset to update the selected model.
- Use model for forecasting. Don’t forget to include prediction intervals.
The helper functions for plotting and the session info are given below.
| |
We can also just use the MLE, e.g.
bxcx(sunspot.year, lambda = 0.417). ↩︎
| Nov 17 | Spectral Analysis | 9 min read |
| Nov 02 | Decomposition and Smoothing Methods | 14 min read |
| Oct 13 | Seasonal Time Series | 17 min read |
| Oct 05 | ARIMA Models | 9 min read |
| Sep 30 | Mean Trend | 13 min read |
 Yi's Knowledge Base
Yi's Knowledge Base